Hello, my name is Beomsoo (Michael) Park ’19 and I am a biomedical engineer at Northwestern University. I am a part of a research group led by Chris Henry focused on developing a new software called KBase, which is a user-friendly program allowing researchers to quickly run genomic analyses to study their own data, rather than relying on other computational scientists to do it for them. KBase allows users to assemble and annotate microbial genomes, build metabolic models, analyze RNA-seq, and overall work with very large quantities of data all at once using “apps” that my Argonne research group designed. KBase has also incorporated numerous other publically available software, such as IDBA, MegaHit, and MetaSPAdes genome assemblies and even quality assessment tools like QUAST. As a new user, my job will be to test KBase to see how compatible and effective the software is. To do this, I will be using KBase to identify unknown species from a large number of soil samples and will eventually write a publication exploring their behaviors by using metabolic modeling and to show off how KBase was used to perform all of the analyses.
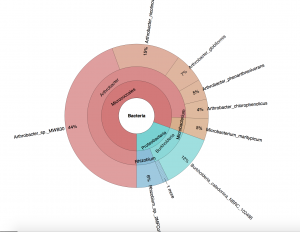
I am currently at the stage in my research where I have fully grasped the understandings behind every program and “app” that I will be using. More specifically, I have been able to upload all of the data onto KBase, run three different types of genome assembly methods (IDBA, MegaHit, MetaSPAdes), and use a program called QUAST to identify the possible species in each particular sample. The pie chart attached shows the distribution of species in one particular sample. For now, I focused on working with a smaller group of samples to use the quality assessment tool within metaQUAST to decide which of the three assembly methods I used is the most accurate, so that I could extrapolate and use this “best method” for all of the samples. In the next coming steps, I will be using KBase to build metabolic models and further exploring the microbial behaviors.
From this summer I wish to learn more about computer programming, since I have a very minimal background knowledge on any programming languages, other than Matlab. I also wish to learn more about metabolic modeling, since I am also very new to this area and was intrigued how we could use computers to predict microbial behaviors. I am hoping to use my time in Argonne in the next following weeks to dive deeper into research to further explore my possible career options.