Developing a High-throughput Functional Bioimaging Capability for Rhizosphere Interactions Utilizing Sensor Cells, Microfluidics, Automation, and AI-guided Analyses
Project Goals: The complex dynamics of root-microbe interactions in the rhizosphere drives recognizable spatial structures. However, knowledge of the specific factors that lead to their development and sustain them for plant health and productivity is sparse.
This project aims to develop a unique functional imaging technique that exploits native sense-and-respond circuits of plant growth-promoting rhizobacteria (PGPR) to monitor chemical exchange between the plant root and microbe during the different phases of colonization. Several native PGPRs are turned into biosensor cells, and root colonization is evaluated with Arabidopsis, camelina, and poplar. Genetic variants of Arabidopsis with gain or loss of function provide drastically altered local environments, resulting in colonization patterns that differ from those observed previously. An orthogonal x-ray imaging approach provides high resolution elemental analysis of the local environment, and imaging throughput in general is accelerated by automation and AI-driven analysis. In addition, we aim to advance the throughput of current bioimaging capabilities that leverage imaging chips developed with BER funding with automation, and an artificial intelligence-guided image analysis strategy.
Isolate Characterization
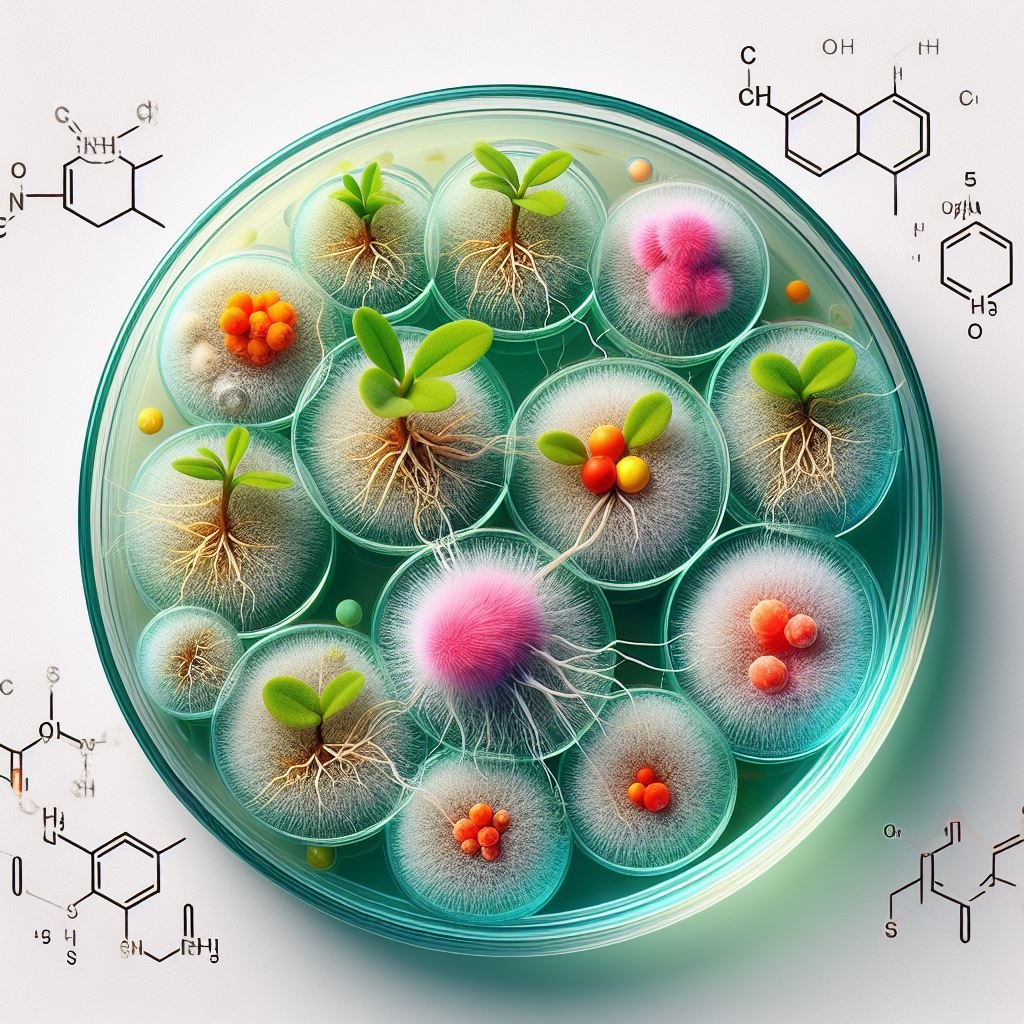
Biosensor Development

AI and Automation

