aggregation selection on Blue Gene
For a lot of workloads, simply using collective I/O provides a big performance boost. Sometimes, though, it’s necessary to tune collective I/O a bit. The hint “cb_nodes” provides a way to select how many MPI processes will become aggregators. On Blue Gene, though, the story is a little more complicated.
We’ll start with Blue Gene /L and /P, even though those machines are now obsolete. The concepts on the older machines still apply, if in a slightly different form. The 163840 cores on the Intrepid BlueGene/P system are configured in a hierarchy. To improve the scalability of the BlueGene architecture, dedicated “I/O nodes” (ION) act as system call proxies between the compute nodes and the storage nodes. On Intrepid, we call the collection of an ION and its compute nodes a “pset”. Each Intrepid pset contains one ION and 64 4-core compute nodes.
The MPI standard defines ‘collective’ routines. Unlike the ‘independent’ routines, all processes in a given MPI communicator call the routine together. The MPI implementation, with the knowledge of which tasks participate in a call, can then perform significant optimizations. These collective routines provide tremendous performance benefits for both networking and I/O.
The BlueGene MPI-IO library, based on ROMIO, makes some adjustments to the ROMIO collective buffering optimization. First, data accesses are aligned to file system block boundaries. Such an alignment reduces lock contention in the write case and can yield big performance improvements.
Second, and perhaps most importantly from a scalability perspective, the “I/O aggregators” selected for the I/O phase of two-phase are a small subset of the total number of processors. On BlueGene, the MPI-IO hint “bgl_nodes_pset” defines a ratio. For each pset allocated to a process, that many nodes will be designated as aggregators. The default ratio for a job running in “virtual node” is one aggregator for every 32 MPI processes. Furthermore, these aggregators are distributed over the topology of the application so that no node has more than one aggregator and no pset contains more than “bgl_nodes_pset” aggregators.
On Mira (Blue Gene /Q) the story is a bit more complicated. I/O nodes no longer are statically assigned to compute nodes. Rather, there is a pool of I/O nodes. When a job is launched, some portion of those I/O nodes gets assigned to the compute nodes.
On Mira, a set of 128 compute nodes (known as a pset) has one I/O node acting as an I/O proxy. For every I/O node there are two network links of 2 GB/s toward two distinct compute nodes acting as bridge. Therefore, for every 128-node partition, there are nb = 1 × 2 = 2 bridges. The I/O traffic from compute nodes passes through these bridge nodes on the way to the I/O node. The I/O nodes are connected to the storage servers through Quad-data-rate (QDR) InfiniBand links. On BG/Q the programmer can set the number of aggregators per pset na_pset (the hint on BG/Q has been renamed to “bg_nodes_pset”). One can determine the total number of aggregators of an application na knowing na_pset , n, and nb with the following equation:
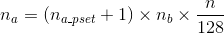
The number of bridge nodes is hardware dependent. For the Argonne machines, Mira’s nb is always 1, but on Vesta, it’s 4 and on Cetus it is 8.
Sophisticated applications wishing to do their own I/O subsetting should be aware of these default parameters and optimizations. In some cases, applications will try to subset to a small number of node and find greatly reduced I/O performance.